|
|
 Run in Google Colab Run in Google Colab
|
 View source on GitHub View source on GitHub
|
Overview
This tutorial demonstrates how to use the embeddings from the Gemini API to detect potential outliers in your dataset. You will visualize a subset of the 20 Newsgroup dataset using t-SNE and detect outliers outside a particular radius of the central point of each categorical cluster.
For more information on getting started with embeddings generated from the Gemini API, check out the Python quickstart.
Prerequisites
You can run this quickstart in Google Colab.
To complete this quickstart on your own development environment, ensure that your envirmonement meets the following requirements:
- Python 3.9+
- An installation of
jupyterto run the notebook.
Setup
First, download and install the Gemini API Python library.
pip install -U -q google.generativeai
import re
import tqdm
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import google.generativeai as genai
import google.ai.generativelanguage as glm
# Used to securely store your API key
from google.colab import userdata
from sklearn.datasets import fetch_20newsgroups
from sklearn.manifold import TSNE
Grab an API Key
Before you can use the Gemini API, you must first obtain an API key. If you don't already have one, create a key with one click in Google AI Studio.
In Colab, add the key to the secrets manager under the "🔑" in the left panel. Give it the name API_KEY.
Once you have the API key, pass it to the SDK. You can do this in two ways:
- Put the key in the
GOOGLE_API_KEYenvironment variable (the SDK will automatically pick it up from there). - Pass the key to
genai.configure(api_key=...)
# Or use `os.getenv('API_KEY')` to fetch an environment variable.
API_KEY=userdata.get('API_KEY')
genai.configure(api_key=API_KEY)
for m in genai.list_models():
if 'embedContent' in m.supported_generation_methods:
print(m.name)
models/embedding-001 models/embedding-001
Prepare dataset
The 20 Newsgroups Text Dataset contains 18,000 newsgroups posts on 20 topics divided into training and test sets. The split between the training and test datasets are based on messages posted before and after a specific date. This tutorial uses the training subset.
newsgroups_train = fetch_20newsgroups(subset='train')
# View list of class names for dataset
newsgroups_train.target_names
['alt.atheism', 'comp.graphics', 'comp.os.ms-windows.misc', 'comp.sys.ibm.pc.hardware', 'comp.sys.mac.hardware', 'comp.windows.x', 'misc.forsale', 'rec.autos', 'rec.motorcycles', 'rec.sport.baseball', 'rec.sport.hockey', 'sci.crypt', 'sci.electronics', 'sci.med', 'sci.space', 'soc.religion.christian', 'talk.politics.guns', 'talk.politics.mideast', 'talk.politics.misc', 'talk.religion.misc']
Here is the first example in the training set.
idx = newsgroups_train.data[0].index('Lines')
print(newsgroups_train.data[0][idx:])
Lines: 15 I was wondering if anyone out there could enlighten me on this car I saw the other day. It was a 2-door sports car, looked to be from the late 60s/ early 70s. It was called a Bricklin. The doors were really small. In addition, the front bumper was separate from the rest of the body. This is all I know. If anyone can tellme a model name, engine specs, years of production, where this car is made, history, or whatever info you have on this funky looking car, please e-mail. Thanks, - IL ---- brought to you by your neighborhood Lerxst ----
# Apply functions to remove names, emails, and extraneous words from data points in newsgroups.data
newsgroups_train.data = [re.sub(r'[\w\.-]+@[\w\.-]+', '', d) for d in newsgroups_train.data] # Remove email
newsgroups_train.data = [re.sub(r"\([^()]*\)", "", d) for d in newsgroups_train.data] # Remove names
newsgroups_train.data = [d.replace("From: ", "") for d in newsgroups_train.data] # Remove "From: "
newsgroups_train.data = [d.replace("\nSubject: ", "") for d in newsgroups_train.data] # Remove "\nSubject: "
# Cut off each text entry after 5,000 characters
newsgroups_train.data = [d[0:5000] if len(d) > 5000 else d for d in newsgroups_train.data]
# Put training points into a dataframe
df_train = pd.DataFrame(newsgroups_train.data, columns=['Text'])
df_train['Label'] = newsgroups_train.target
# Match label to target name index
df_train['Class Name'] = df_train['Label'].map(newsgroups_train.target_names.__getitem__)
df_train
Next, sample some of the data by taking 150 data points in the training dataset and choosing a few categories. This tutorial uses the science categories.
# Take a sample of each label category from df_train
SAMPLE_SIZE = 150
df_train = (df_train.groupby('Label', as_index = False)
.apply(lambda x: x.sample(SAMPLE_SIZE))
.reset_index(drop=True))
# Choose categories about science
df_train = df_train[df_train['Class Name'].str.contains('sci')]
# Reset the index
df_train = df_train.reset_index()
df_train
df_train['Class Name'].value_counts()
sci.crypt 150 sci.electronics 150 sci.med 150 sci.space 150 Name: Class Name, dtype: int64
Create the embeddings
In this section, you will see how to generate embeddings for the different texts in the dataframe using the embeddings from the Gemini API.
API changes to Embeddings with model embedding-001
For the new embeddings model, embedding-001, there is a new task type parameter and the optional title (only valid with task_type=RETRIEVAL_DOCUMENT).
These new parameters apply only to the newest embeddings models.The task types are:
| Task Type | Description |
|---|---|
| RETRIEVAL_QUERY | Specifies the given text is a query in a search/retrieval setting. |
| RETRIEVAL_DOCUMENT | Specifies the given text is a document in a search/retrieval setting. |
| SEMANTIC_SIMILARITY | Specifies the given text will be used for Semantic Textual Similarity (STS). |
| CLASSIFICATION | Specifies that the embeddings will be used for classification. |
| CLUSTERING | Specifies that the embeddings will be used for clustering. |
from tqdm.auto import tqdm
tqdm.pandas()
from google.api_core import retry
def make_embed_text_fn(model):
@retry.Retry(timeout=300.0)
def embed_fn(text: str) -> list[float]:
# Set the task_type to CLUSTERING.
embedding = genai.embed_content(model=model,
content=text,
task_type="clustering")['embedding']
return np.array(embedding)
return embed_fn
def create_embeddings(df):
model = 'models/embedding-001'
df['Embeddings'] = df['Text'].progress_apply(make_embed_text_fn(model))
return df
df_train = create_embeddings(df_train)
df_train.drop('index', axis=1, inplace=True)
0%| | 0/600 [00:00<?, ?it/s]
Dimensionality reduction
The dimension of the document embedding vector is 768. In order to visualize how the embedded documents are grouped together, you will need to apply dimensionality reduction as you can only visualize the embeddings in 2D or 3D space. Contextually similar documents should be closer together in space as opposed to documents that are not as similar.
len(df_train['Embeddings'][0])
768
# Convert df_train['Embeddings'] Pandas series to a np.array of float32
X = np.array(df_train['Embeddings'].to_list(), dtype=np.float32)
X.shape
(600, 768)
You will apply the t-Distributed Stochastic Neighbor Embedding (t-SNE) approach to perform dimensionality reduction. This technique reduces the number of dimensions, while preserving clusters (points that are close together stay close together). For the original data, the model tries to construct a distribution over which other data points are "neighbors" (e.g., they share a similar meaning). It then optimizes an objective function to keep a similar distribution in the visualization.
tsne = TSNE(random_state=0, n_iter=1000)
tsne_results = tsne.fit_transform(X)
df_tsne = pd.DataFrame(tsne_results, columns=['TSNE1', 'TSNE2'])
df_tsne['Class Name'] = df_train['Class Name'] # Add labels column from df_train to df_tsne
df_tsne
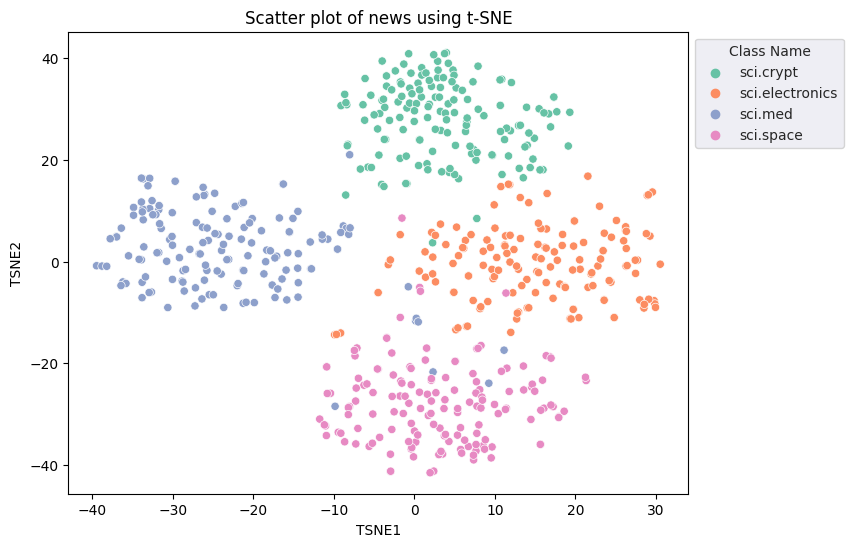
fig, ax = plt.subplots(figsize=(8,6)) # Set figsize
sns.set_style('darkgrid', {"grid.color": ".6", "grid.linestyle": ":"})
sns.scatterplot(data=df_tsne, x='TSNE1', y='TSNE2', hue='Class Name', palette='Set2')
sns.move_legend(ax, "upper left", bbox_to_anchor=(1, 1))
plt.title('Scatter plot of news using t-SNE')
plt.xlabel('TSNE1')
plt.ylabel('TSNE2');
Outlier detection
To determine which points are anomalous, you will determine which points are inliers and outliers. Start by finding the centroid, or location that represents the center of the cluster, and use the distance to determine the points that are outliers.
Start by getting the centroid of each category.
def get_centroids(df_tsne):
# Get the centroid of each cluster
centroids = df_tsne.groupby('Class Name').mean()
return centroids
centroids = get_centroids(df_tsne)
centroids
def get_embedding_centroids(df):
emb_centroids = dict()
grouped = df.groupby('Class Name')
for c in grouped.groups:
sub_df = grouped.get_group(c)
# Get the centroid value of dimension 768
emb_centroids[c] = np.mean(sub_df['Embeddings'], axis=0)
return emb_centroids
emb_c = get_embedding_centroids(df_train)
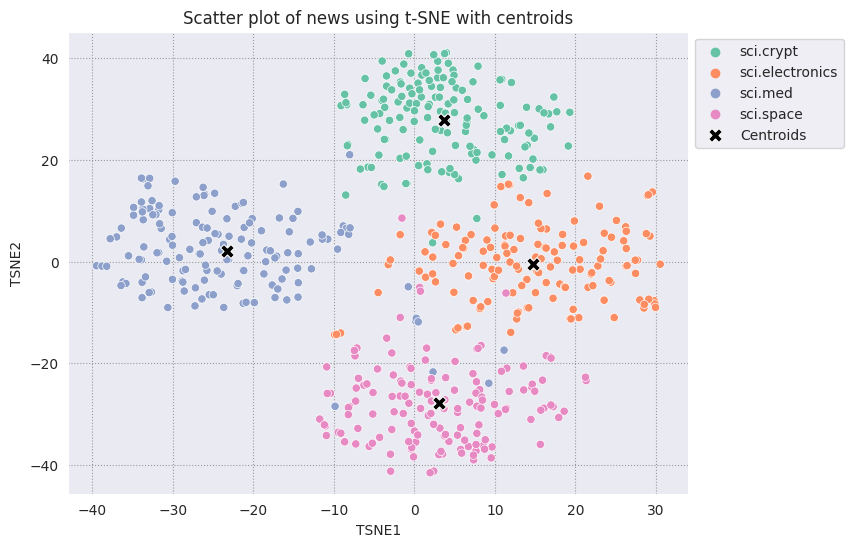
Plot each centroid you have found against the rest of the points.
# Plot the centroids against the cluster
fig, ax = plt.subplots(figsize=(8,6)) # Set figsize
sns.set_style('darkgrid', {"grid.color": ".6", "grid.linestyle": ":"})
sns.scatterplot(data=df_tsne, x='TSNE1', y='TSNE2', hue='Class Name', palette='Set2');
sns.scatterplot(data=centroids, x='TSNE1', y='TSNE2', color="black", marker='X', s=100, label='Centroids')
sns.move_legend(ax, "upper left", bbox_to_anchor=(1, 1))
plt.title('Scatter plot of news using t-SNE with centroids')
plt.xlabel('TSNE1')
plt.ylabel('TSNE2');
Choose a radius. Anything beyond this bound from the centroid of that category is considered an outlier.
def calculate_euclidean_distance(p1, p2):
return np.sqrt(np.sum(np.square(p1 - p2)))
def detect_outlier(df, emb_centroids, radius):
for idx, row in df.iterrows():
class_name = row['Class Name'] # Get class name of row
# Compare centroid distances
dist = calculate_euclidean_distance(row['Embeddings'],
emb_centroids[class_name])
df.at[idx, 'Outlier'] = dist > radius
return len(df[df['Outlier'] == True])
range_ = np.arange(0.3, 0.75, 0.02).round(decimals=2).tolist()
num_outliers = []
for i in range_:
num_outliers.append(detect_outlier(df_train, emb_c, i))
# Plot range_ and num_outliers
fig = plt.figure(figsize = (14, 8))
plt.rcParams.update({'font.size': 12})
plt.bar(list(map(str, range_)), num_outliers)
plt.title("Number of outliers vs. distance of points from centroid")
plt.xlabel("Distance")
plt.ylabel("Number of outliers")
for i in range(len(range_)):
plt.text(i, num_outliers[i], num_outliers[i], ha = 'center')
plt.show()
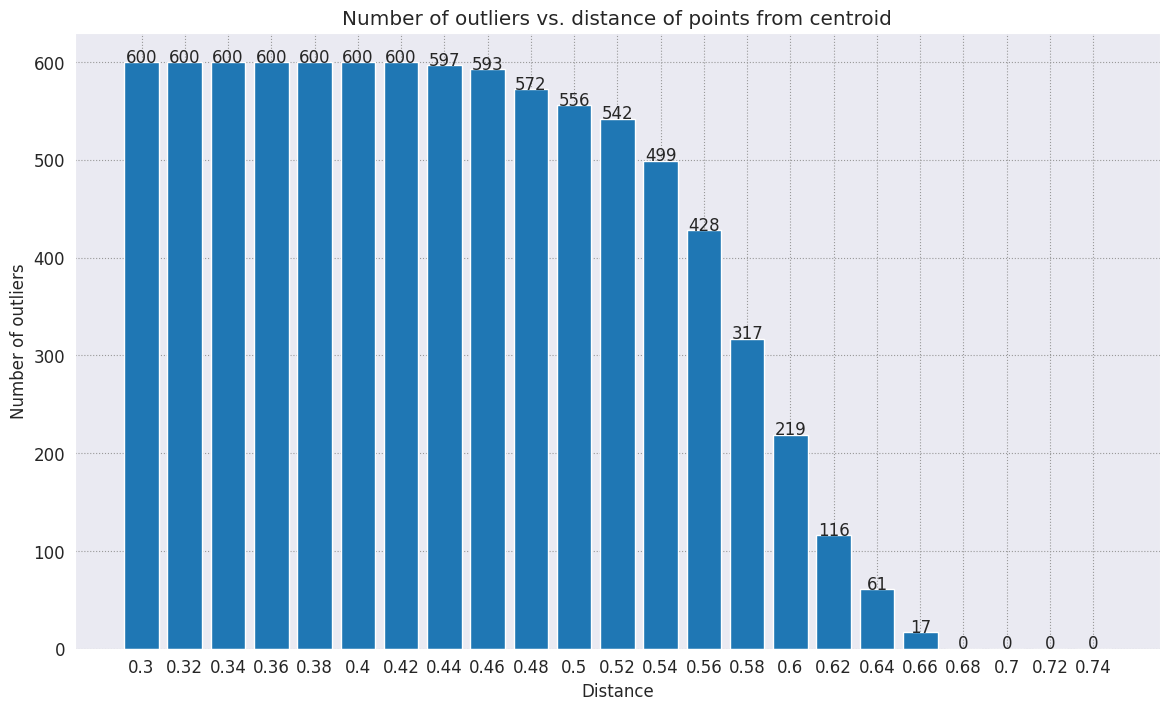
Depending on how sensitive you want your anomaly detector to be, you can choose which radius you would like to use. For now, 0.62 is used, but you can change this value.
# View the points that are outliers
RADIUS = 0.62
detect_outlier(df_train, emb_c, RADIUS)
df_outliers = df_train[df_train['Outlier'] == True]
df_outliers.head()
# Use the index to map the outlier points back to the projected TSNE points
outliers_projected = df_tsne.loc[df_outliers['Outlier'].index]
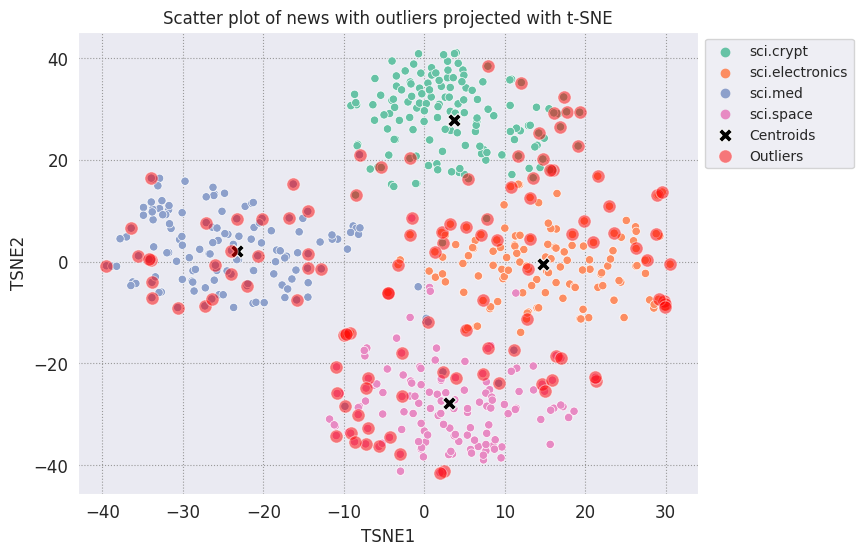
Plot the outliers and denote them using a transparent red color.
fig, ax = plt.subplots(figsize=(8,6)) # Set figsize
plt.rcParams.update({'font.size': 10})
sns.set_style('darkgrid', {"grid.color": ".6", "grid.linestyle": ":"})
sns.scatterplot(data=df_tsne, x='TSNE1', y='TSNE2', hue='Class Name', palette='Set2');
sns.scatterplot(data=centroids, x='TSNE1', y='TSNE2', color="black", marker='X', s=100, label='Centroids')
# Draw a red circle around the outliers
sns.scatterplot(data=outliers_projected, x='TSNE1', y='TSNE2', color='red', marker='o', alpha=0.5, s=90, label='Outliers')
sns.move_legend(ax, "upper left", bbox_to_anchor=(1, 1))
plt.title('Scatter plot of news with outliers projected with t-SNE')
plt.xlabel('TSNE1')
plt.ylabel('TSNE2');
Use the index values of the datafames to print a few examples of what outliers can look like in each category. Here, the first data point from each category is printed out. Explore other points in each category to see data that are deemed as outliers, or anomalies.
sci_crypt_outliers = df_outliers[df_outliers['Class Name'] == 'sci.crypt']
print(sci_crypt_outliers['Text'].iloc[0])
Re: Source of random bits on a Unix workstation Lines: 44 Nntp-Posting-Host: sandstorm >>For your application, what you can do is to encrypt the real-time clock >>value with a secret key. Well, almost.... If I only had to solve the problem for myself, and were willing to have to type in a second password whenever I logged in, it could work. However, I'm trying to create a solution that anyone can use, and which, once installed, is just as effortless to start up as the non-solution of just using xhost to control access. I've got religeous problems with storing secret keys on multiuser computers. >For a good discussion of cryptographically "good" random number >generators, check out the draft-ietf-security-randomness-00.txt >Internet Draft, available at your local friendly internet drafts >repository. Thanks for the pointer! It was good reading, and I liked the idea of using several unrelated sources with a strong mixing function. However, unless I missed something, the only source they suggested that seems available, and unguessable by an intruder, when a Unix is fresh-booted, is I/O buffers related to network traffic. I believe my solution basically uses that strategy, without requiring me to reach into the kernel. >A reasonably source of randomness is the output of a cryptographic >hash function , when fed with a large amount of >more-or-less random data. For example, running MD5 on /dev/mem is a >slow, but random enough, source of random bits; there are bound to be >128 bits of entropy in the tens of megabytes of data in >a modern workstation's memory, as a fair amount of them are system >timers, i/o buffers, etc. I heard about this solution, and it sounded good. Then I heard that folks were experiencing times of 30-60 seconds to run this, on reasonably-configured workstations. I'm not willing to add that much delay to someone's login process. My approach takes a second or two to run. I'm considering writing the be-all and end-all of solutions, that launches the MD5, and simultaneously tries to suck bits off the net, and if the net should be sitting __SO__ idle that it can't get 10K after compression before MD5 finishes, use the MD5. This way I could have guaranteed good bits, and a deterministic upper bound on login time, and still have the common case of login take only a couple of extra seconds. -Bennett
sci_elec_outliers = df_outliers[df_outliers['Class Name'] == 'sci.electronics']
print(sci_elec_outliers['Text'].iloc[0])
Re: Laser vs Bubblejet?
Reply-To:
Distribution: world
X-Mailer: cppnews \\(Revision: 1.20 \\)
Organization: null
Lines: 53
Here is a different viewpoint.
> FYI: The actual horizontal dot placement resoution of an HP
> deskjet is 1/600th inch. The electronics and dynamics of the ink
> cartridge, however, limit you to generating dots at 300 per inch.
> On almost any paper, the ink wicks more than 1/300th inch anyway.
>
> The method of depositing and fusing toner of a laster printer
> results in much less spread than ink drop technology.
In practice there is little difference in quality but more care is needed
with inkjet because smudges etc. can happen.
> It doesn't take much investigation to see that the mechanical and
> electronic complement of a laser printer is more complex than
> inexpensive ink jet printers. Recall also that laser printers
> offer a much higher throughput: 10 ppm for a laser versus about 1
> ppm for an ink jet printer.
A cheap laser printer does not manage that sort of throughput and on top of
that how long does the _first_ sheet take to print? Inkjets are faster than
you say and in both cases the computer often has trouble keeping up with the
printer.
A sage said to me: "Do you want one copy or lots of copies?", "One",
"Inkjet".
> Something else to think about is the cost of consumables over the
> life of the printer. A 3000 page yield toner cartridge is about
> $US 75-80 at discount while HP high capacity
> cartridges are about $US 22 at discount. It could be that over the
> life cycle of the printer that consumables for laser printers are
> less than ink jet printers. It is getting progressively closer
> between the two technologies. Laser printers are usually desinged
> for higher duty cycles in pages per month and longer product
> replacement cycles.
Paper cost is the same and both can use refills. Long term the laserprinter
will need some expensive replacement parts and on top of that
are the amortisation costs which favour the lowest purchase cost printer.
HP inkjets understand PCL so in many cases a laserjet driver will work if the
software package has no inkjet driver.
There is one wild difference between the two printers: a laserprinter is a
page printer whilst an inkjet is a line printer. This means that a
laserprinter can rotate graphic images whilst an inkjet cannot. Few drivers
actually use this facility.
TC.
E-mail: or
sci_med_outliers = df_outliers[df_outliers['Class Name'] == 'sci.med']
print(sci_med_outliers['Text'].iloc[0])
Re: THE BACK MACHINE - Update Organization: University of Nebraska--Lincoln Lines: 15 Distribution: na NNTP-Posting-Host: unlinfo.unl.edu I have a BACK MACHINE and have had one since January. While I have not found it to be a panacea for my back pain, I think it has helped somewhat. It MAINLY acts to stretch muscles in the back and prevent spasms associated with pain. I am taking less pain medication than I was previously. The folks at BACK TECHNOLOGIES are VERY reluctant to honor their return policy. They extended my "warranty" period rather than allow me to return the machine when, after the first month or so, I was not thrilled with it. They encouraged me to continue to use it, abeit less vigourously. Like I said, I can't say it is a cure-all, but it keeps me stretched out and I am in less pain. -- *********************************************************************** Dale M. Webb, DVM, PhD * 97% of the body is water. The Veterinary Diagnostic Center * other 3% keeps you from drowning. University of Nebraska, Lincoln *
sci_space_outliers = df_outliers[df_outliers['Class Name'] == 'sci.space']
print(sci_space_outliers['Text'].iloc[0])
MACH 25 landing site bases? Article-I.D.: aurora.1993Apr5.193829.1 Organization: University of Alaska Fairbanks Lines: 7 Nntp-Posting-Host: acad3.alaska.edu The supersonic booms hear a few months ago over I belive San Fran, heading east of what I heard, some new super speed Mach 25 aircraft?? What military based int he direction of flight are there that could handle a Mach 25aircraft on its landing decent?? Odd question?? == Michael Adams, -- I'm not high, just jacked
Next steps
You've now created an anomaly detector using embeddings! Try using your own textual data to visualize them as embeddings, and choose some bound such that you can detect outliers. You can perform dimensionality reduction in order to complete the visualization step. Note that t-SNE is good at clustering inputs, but can take a longer time to converge or might get stuck at local minima. If you run into this issue, another technique you could consider are principal components analysis (PCA).
To learn how to use other services in the Gemini API, visit the Python quickstart.
To learn more about how you can use the embeddings, check out the examples available. To learn how to create them from scratch, see TensorFlow's Word Embeddings tutorial.
